Its been a while since i have written to this post. I have been fiddling around with VTK. Its quite hell of a learning curve when you are teaching VTK to yourself. Here are few things I have to say about VTK:
1) Its amazing that the visualization library is open source.
2) You will hate VTK from the day you start learning it, till the day you master it. It's the most wonderful thing when you have mastered it.
3) It's a nightmare if you can't debug your VTK programs, make sure your debugger works with VTK
4) Make sure you know how to look into VTK documentation. The documentation has been
generated by DOxygen, and make sure you know how to find functions, how to list them alphabetically, etc. It really helps if you can know how to quickly search through documentation. Generally, I look through 'VTK Class members' (see here ), class members are functions ofcourse. There is an alphabetic index on top of this page, so if you are searching for 'foo', click on f and search through page.
5) The best way to solve your VTK problems is by going through the VTK forums. Their search utility is not the greatest, but you can search intelligently, once you know how their search engine behaves. Here's the VTK forum.
6) ... and the bottom line: if you are not an avid C++ programmer, you will be surprised at how small things can cause segmentation faults within a VTK program (make sure you have called your constructors, etc). Have patience, once you know how VTK behaves, you will enjoy using the VTK library.
7) .. and hey .. dont forget to get a dedicated graphics card for your PC (not the ones which come with the motherboard). It really helps with your renderings!
My PhD progress is quite good, these days I am trying to create an interactive segmentation tool using VTK + Imperial college's ITK + FLTK. I had trouble trying to embed VTK windows into FLTK GUIs, and I was successfully able to do it. There are third pirty libraries out there such as the vtkFltk library (just 2 files), which is the most easiest to use.
Apart from this I am able to view the MRI volume using two orthogonal planes (which slice through the volume), here are orthogonal views of a brain MRI:

And here's the hypothalamus segmented by allowing the user to specify a region of interest:
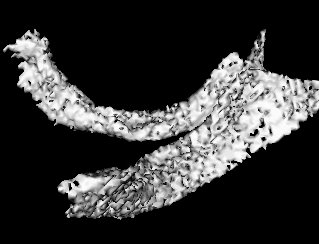
These were segmented using a region-growing approach with a simple upper-lower threshold technique. This requires a great deal of trial and error to choose the correct upper and lower threshold limits.
1) Its amazing that the visualization library is open source.
2) You will hate VTK from the day you start learning it, till the day you master it. It's the most wonderful thing when you have mastered it.
3) It's a nightmare if you can't debug your VTK programs, make sure your debugger works with VTK
4) Make sure you know how to look into VTK documentation. The documentation has been
generated by DOxygen, and make sure you know how to find functions, how to list them alphabetically, etc. It really helps if you can know how to quickly search through documentation. Generally, I look through 'VTK Class members' (see here ), class members are functions ofcourse. There is an alphabetic index on top of this page, so if you are searching for 'foo', click on f and search through page.
5) The best way to solve your VTK problems is by going through the VTK forums. Their search utility is not the greatest, but you can search intelligently, once you know how their search engine behaves. Here's the VTK forum.
6) ... and the bottom line: if you are not an avid C++ programmer, you will be surprised at how small things can cause segmentation faults within a VTK program (make sure you have called your constructors, etc). Have patience, once you know how VTK behaves, you will enjoy using the VTK library.
7) .. and hey .. dont forget to get a dedicated graphics card for your PC (not the ones which come with the motherboard). It really helps with your renderings!
My PhD progress is quite good, these days I am trying to create an interactive segmentation tool using VTK + Imperial college's ITK + FLTK. I had trouble trying to embed VTK windows into FLTK GUIs, and I was successfully able to do it. There are third pirty libraries out there such as the vtkFltk library (just 2 files), which is the most easiest to use.
Apart from this I am able to view the MRI volume using two orthogonal planes (which slice through the volume), here are orthogonal views of a brain MRI:

And here's the hypothalamus segmented by allowing the user to specify a region of interest:

These were segmented using a region-growing approach with a simple upper-lower threshold technique. This requires a great deal of trial and error to choose the correct upper and lower threshold limits.
No comments:
Post a Comment