After spending an entire week optimizing and debugging code, I am finally able to segment the left atrium semi-automatically. Semi-automatic in the sense that it requires user-interaction where the user first needs to crop a region-grown segmented MRI, approximately locating where the atrium lies. This step is not essential to the algorithm, however, it speeds up significantly the EDT (Euclidean distance transform) computation. In the next step, the local maximum points are computed. Interestingly, these were arranged in a very interesting fashion on the atrium. I will discuss this in further detail on future posts.
After computing the local maximas, for each voxel in the segmented MRI, its basic component is computed. A voxel's basic component is its closest reachable local maxima where the path must be the one with increasing EDT values. I have noticed that some voxel's don't belong to a basic component. I call these voxels orphans. These usually occur when during a basic component path search, the search-path explodes into a neighborhood of voxels with zero EDT values. In which case, there is no one local maxima point and hence no basic component. These may cause discontinuity within an atrium or its drainage vessels.
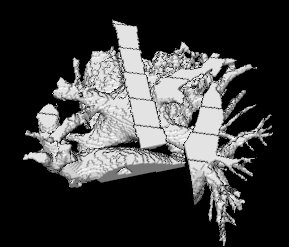
Here is the original image, from which the atrium was segmented, one can hardly see the atrium as it is surrounded by a dense mesh of blood vessels and the aorta.
And here's the segmented atrium:
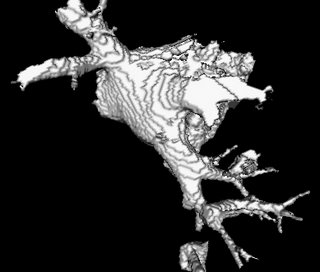
In the final segmentation step, the user clicks on local-maximums which approximately lie inside the atrium. This reveals all voxels which belong to basic components centered by the selected local-maximas. This system cant be navigated by someone who hasn't seen an atrium before.
The segmentation technique is quite powerful and it can prove to be very useful in extracting structures that are embedded within a dense mesh of blood-vessel-like structures like the atrium.
Programming note:
func (a)
{
for ( .. )
*p = new someClass(a,b,c)
}
Doing the above, for some reason, doesn't clear up memory held by someClass objects on exiting the function func.
Here is the original image, from which the atrium was segmented, one can hardly see the atrium as it is surrounded by a dense mesh of blood vessels and the aorta.

And here's the segmented atrium:

In the final segmentation step, the user clicks on local-maximums which approximately lie inside the atrium. This reveals all voxels which belong to basic components centered by the selected local-maximas. This system cant be navigated by someone who hasn't seen an atrium before.
The segmentation technique is quite powerful and it can prove to be very useful in extracting structures that are embedded within a dense mesh of blood-vessel-like structures like the atrium.
Programming note:
func (a)
{
for ( .. )
*p = new someClass(a,b,c)
}
Doing the above, for some reason, doesn't clear up memory held by someClass objects on exiting the function func.
1 comment:
Good work buddy!!!!! Seems u really into it now...keep it up....
Ur MSc. Classmate @ QMUL
Nitish Anand
www.freewebs.com/nitish-anand
Post a Comment